
RESEARCH HIGHLIGHTS

Development of first generation in-situ pathogen detection system (Gen1-IPDS) based on NanoGene assay for near real time E. coli O157:H7 detection
We developed the first generation in-situ pathogen detection system (Gen1-IPDS) based on the NanoGene assay for detecting and quantifying Escherichia coli O157:H7 specific eaeA gene. The Gen1-IPDS is currently capable of executing four key steps required in the NanoGene assay: sample and reagents introduction, DNA hybridization, magnetic separation of complexes, and sample collection. Operational parameters such as magnet position, hybridization buffer composition, hybridization flow rate, and hybridization temperature were investigated. Using the experimentally determined operational parameters, the target gene was successfully quantified (R2=0.97) over a range of six orders of magnitude (10−12 to 10−6 mol L−1). The NanoGene assay quantification results via Gen1-IPDS were validated by correlation with its laboratory version (R2=0.97).
Mitchell, K.; Chua, B.; *Son, A. (2014) Biosensors and Bioelectronics, 54, 229-236.
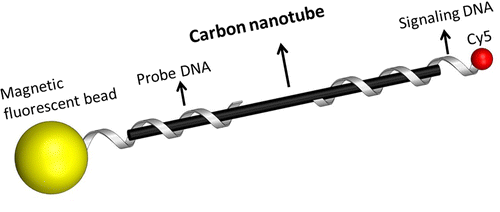

Quantitative detecton of single walled carbon nanotube in water using DNA and magnetic fluorescent spheres
Carbon nanotubes (CNTs) possess unique properties that have led to an increase in their research and usage for a wide variety of fields. This growing demand of CNTs poses a major public health risk given its unregulated release into the environment. Unfortunately there is a significant information gap on the actual quantity of CNTs in the environment due to limitation of existing detection methods. This is mainly owing to the ubiquitous carbon chemistry of CNT. In response we developed a method (CNT-capture method) that is able to structurally differentiate CNT from other interference carbon materials in an aqueous medium. The affinity between single walled nanotubes (SWNTs) and specific single stranded DNA (ssDNA) was employed to capture SWNTs in water. SWNT-specific separation was obtained via magnetic separation. Dual fluorescent labels attached to sandwich ssDNA probes were used for quantification. With optimized parameters, a calibration curve of quantification (R2 = 0.90) was obtained with a range of SWNT concentration (0.05–10 μg/mL) against graphene as a planar analog. Comparison to other spectroscopy based methods was carried out to highlight the specificity and sensitivity of the presented method for CNT detection in aquatic sample.
Mota, L.C.; Urena-Benavides, E.E.; Yoon, Y.; *Son, A. (2013) Environmental Science and Technology, 47, 493-501.



Quantification of E. coli O157:H7 using an inhibitor resistant NanoGene assay
Humic acids are ubiquitous and abundant in terrestrial environments; therefore, they are often co-extracted with nucleic acids and interfere with quantitative PCR (qPCR) assays. In this study a recently developed NanoGene assay that is resistant to interference by humic acids was evaluated for gene detection in soil samples. The NanoGene assay utilizes a combination of magnetic beads, dual quantum dots labels, and DNA hybridization in solution. Seven soil samples containing different amounts of organic matter were tested to compare NanoGene and qPCR assays for their respective ability to detect a bacterial pathogen. We spiked the soils with Escherichia coli O157:H7, extracted genomic DNA, and conducted NanoGene and qPCR assays targeting the E. coli O157:H7-specific eaeA gene. Compared to the qPCR assay the NanoGene assay was significantly more resistant to the inhibitory effect of humic acids, successfully quantifying the eaeA gene within a linear (R2 = 0.99) range of 10^5 through 10^8 CFU/g soil for all seven soil samples tested. In contrast, the qPCR assay was significantly inhibited using the same template DNA isolated from soils containing a range of organic content (2.0%–12%). This study demonstrated that the NanoGene assay is suitable for quantitative gene detection in diverse soil types and is not susceptible to inhibition by humic acids and other organic compounds that commonly lead to false negative results in qPCR assays.
Wang, X.; Liles, M.R.; *Son, A. (2013) Soil Biology and Biochemistry, 58, 9-15.
Kim, G.; Wang, X.; Ahn, H.; *Son, A. (2011) Environmental Science and Technology, 45, 8873-8880.
Kim, G.; Wang, X.; *Son, A. (2011) J. Environmental Monitoring, 13, 1344-1350.
Kim, G.; Son, A. (2010) Analytica Chimica Acta, 677, 90-96.




Microbial community analysis of Deepwater Horizon oil-spill impacted sites along the Gulf coast using functional and phylogenetic markers
We investigated the impact of the Deepwater Horizon oil spill on microbial communities in wetland sediment and seawater samples collected from sites along the Gulf shore. Based on GC/MS analysis, the sediment from Bay Jimmy, LA had detectable signs of hydrocarbon contamination, identified as n-alkanes in the GC/MS spectrum similar to that of the Deepwater Horizon source oil (MC-252). To identify changes in microbial assemblage structure and functional diversity in response to hydrocarbon contamination, five genes (bacterial 16S rRNA, Pseudomonas-specific 16S rRNA, alkB, P450, and PAH-RHDα) were selected based on the specific enzymes encoded by bacteria to degrade alkanes or polycyclic aromatic hydrocarbons. A ribotype analysis based on pyrosequencing identified 17 bacteria genera known for their capacity to degrade hydrocarbons, including Mycobacterium, Novosphingobium, Parvibaculum, Pseudomonas, and Sphingomonas, in the contaminated sediment sample. Furthermore, the contaminated sample had a very high relative abundance of 16S rRNA gene sequences affiliated with the genus Parvibaculum, members of which have been characterized for their degradative abilities. These data suggest that specific bacterial taxa within the genus Parvibaculum have the capacity for hydrocarbon degradation and could use the hydrocarbons as a carbon and energy source, resulting in a dominant population in a hydrocarbon-contaminated soil. In summary, when exposed to the spilled oil, the distinct wetland microbial communities responded with decreased diversity and increased abundance of selective degradative species.
Looper, J.; Cotto, A.; Kim, B.-Y.; Lee, M.-K.; Ni Chadhain, S.; Liles, M.; *Son, A. (2014) Environmental Science: Processes & Impacts 15, 2068-2079.
Natter, M.; Keevan, J.; Wang, Y.; Keimowitz, A.S.; Okeke, B.; Son, A.; Lee, M. (2012) Environmental Science and Technology, 47, 493-501.


Physical Lysis Only (PLO) methods suitable as rapid sample pretreatment for qPCR and NanoGene assay
Quantitative PCR enables rapid and sensitive gene quantification and is widely used in genomics, such as biological, medical, environmental, and food sciences. However sample pretreatment requires the use of conventional DNA extraction kits which are time consuming and labor intensive. We investigated four physical lysis only (PLO) methods which are rapid and could serve as alternatives to conventional DNA extraction kits. These PLO methods are bead mill, heating, sonication, and freeze-thaw. Using ethidium bromide based assay, their performance were evaluated and compared. The effects of cell debris and its removal were also investigated. Bead mill method without cell debris removal appeared to yield the best qPCR results among the four PLO methods. In addition, bead mill method also performed better than conventional DNA extraction kits. It is probably due to the substantial loss of DNA material during the extensive purification of the conventional DNA extraction kits. The bead mill method has been demonstrated to successfully quantify 10^2 to 10^7 copies of the PAH-RHDα gene of Pseudomonas putida.
Wang, X.; *Son, A. (2013) Environmental Science: Processes & Impacts, 15, 2068-2079.
Wang, X.; Lim, H.; *Son, A. (2014) Environmental Health & Toxicology (In print).
Wang, X.; Lee, B.; *Son, A. (2014) Applied Microbiology and Biotechnology (In print)
Wang, X.; *Son, A. Environmental Science and Technology (Under review)